Pearman JK, Sissons J, Kihika JK, Thomson-Laing G, Wood SA (2025) Microbial biodiversity and metabolic functioning in sediments of coastal dune lakes on a remote island. Metabarcoding and Metagenomics 9: e144128. https://doi.org/10.3897/mbmg.9.144128
Abstract
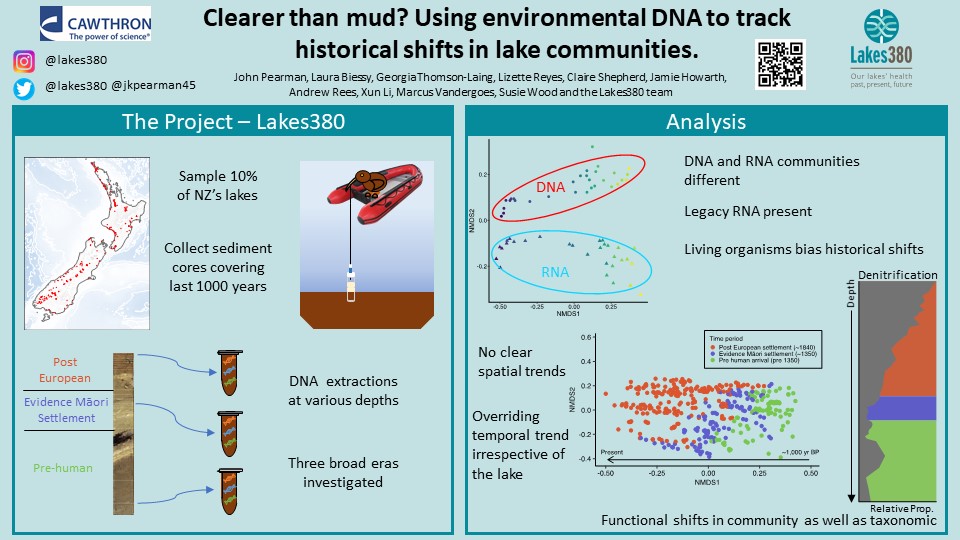
Molecular-based techniques provide the potential for novel insights into the functioning of ecosystems, especially those that are globally rare such as coastal dune lakes. In the surface sediments of lakes, microbial communities play a vital role in biogeochemical cycling and techniques such as metagenomics can provide information on the roles play in these ecosystems. The current study aimed to investigate the taxonomic and functional composition of six coastal dune lakes on Chatham Island using sediment DNA approaches. The use of metabarcoding (16S rRNA gene) and metagenomics showed that there were distinct differences in the microbial community composition and functional potential amongst the lakes, especially in the lakes with higher salinity. Investigation of metabolic potential with metagenomics showed that the abundance of genes involved in nitrogen cycling were related to the nitrogen:phosphorus ratio while assimilatory sulfate reduction was correlated with sulfur and organic matter concentrations. Analysis showed differences in the carbon fixation strategies amongst the lakes. The lake with the highest salinity levels also had elevated levels of osmoprotectants and related transporters. The sequencing of sediment DNA enables the investigation of the composition and functioning of lake environments providing a basis for the increased understanding of the processes occurring within lakes.